Project 09
Artificial intelligence-guided molecular dynamics simulations decipher the activation mechanism of cellular control programs
Principle Investigator: Dr. Roberto Covino
Research Areas: Biophysics, Statistical Physics, Biological Physics
Summary
In cellular membranes, lipids and proteins self-assemble into heterogeneous and dynamic complexes. Membrane proteins and their environment are engaged in a constant dialogue, which is at the origin of many essential regulatory phenomena. Despite its importance, studying the assembly mechanism of membrane proteins with standard approaches is incredibly challenging. We will integrate membrane and protein biophysics, multi-scale molecular dynamics simulations, statistical mechanics, and artificial intelligence to reveal the assembly mechanism in atomistic detail. Modern path theory will provide the thermodynamics and kinetics of the process, which will allow us to understand the role of generic bilayer- mediated interactions. The unfolded protein response (UPR) is a fundamental eukaryotic homeostatic mechanism that protects the cell from stresses threatening its survival. The UPR is at the center of a cellular life or death decision. The ER-transmembrane protein Ire1 controls the activation of the most conserved branch of the UPR. When Ire1 senses ER-stress, it assembles into dimers, oligomers, and large clusters, that initiate a transcriptional and translational response. Despite their fundamental role, the early events determining the UPR’s activation are poorly understood. Integrating experimental and theoretical methods, we will dissect the structural dynamics of the human Ire1 and its organization in dimers and larger supramolecular assemblies. Given the growing interest in pharmacological targeting of Ire1, a deep mechanistic understanding of the UPR’s activation is of outstanding biological and biomedical importance.
P09: PROJECT-RELATED PUBLICATIONS
- Spinetti E, Karagöz GE, Covino R* (2025) The structural dynamics of IRE1 and its interaction with unfolded peptides. eLife 14: RP106716
- Lazzeri G, Bolhuis PG, Covino R* (2025) Optimal rejection-free path sampling. arXiv preprint: 2503.21037
- Jackel E, Lazzeri G, Covino R* (2025) Free energy, rates, and mechanism of transmembrane dimerization in lipid bilayers from dynamically unbiased molecular dynamics simulations. J Phys Chem B 129: 1586-1596
- Kettel P, Marosits L, Spinetti E, Rechberger M, Giannini C, Radler P, Niedermoser I, Fischer I, Versteeg GA, Loose M, Covino R, Karagoz GE* (2024) Disordered regions in the IRE1α ER luminal domain mediate its stress-induced clustering. EMBO J 43: 4668-4698
- Li Y, Arghittu SM, Dietz MS, Hella GJ, Haße D, Ferraris DM, Freund P, Barth H, Iamele L, de Jonge H, Niemann HH, Covino R*, Heilemann M* (2024) Single-molecule imaging and molecular dynamics simulations reveal early activation of the MET receptor in cells. Nat Commun 15: 9486
- Lazzeri G, Jung H, Bolhuis PG, Covino R* (2023) Molecular free energies, rates, and mechanisms from data-efficient path sampling simulations. J Chem Theory Comput 19: 9060-9076
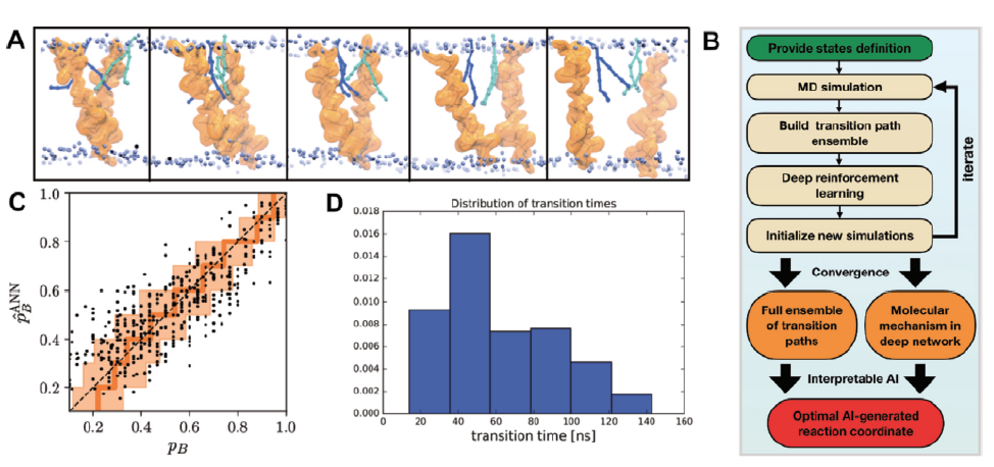
- Jung H#, Covino R#, Arjun A, Leitlold C, Dell’ago C, Bolhuis P, Hummer G* (2023) Machine-guided path sampling to discover mechanisms of molecular self-organization. Nat Comput Sci 3: 334-345
- Nishimura T, Lazzeri G, Mizushima N, Covino R and Tooze SA (2023) Unique amphipathic a helix drives membrane insertion and enzymatic activity of ATG3. Sci Adv 9: eadh1281
- Dingeldein L, Cossio P and Covino R (2023) Simulation-based inference of single-molecule force spectroscopy. Sci Technol 4: 025009
- Turoňová B, Sikora M, Schürmann C, Hagen WJH., Welsch S., Blanc FEC., Bülow Sv, Gecht M, Bagola K, Hörner C, Zandbergen Gv, Landry J, Azevedo NTDd, Mosalaganti S, Schwarz A, Covino R, Mühlebach MD, Hummer G, Krijnse Locker J, and Beck M (2020) In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 370: 203

