Project 02
Mechanism of bacterial transenvelope protein and lipid transport
Principle Investigator: Prof. Dr. Benesh Joseph
Research Areas: Membrane Biophysics, EPR Spectroscopy
Summary
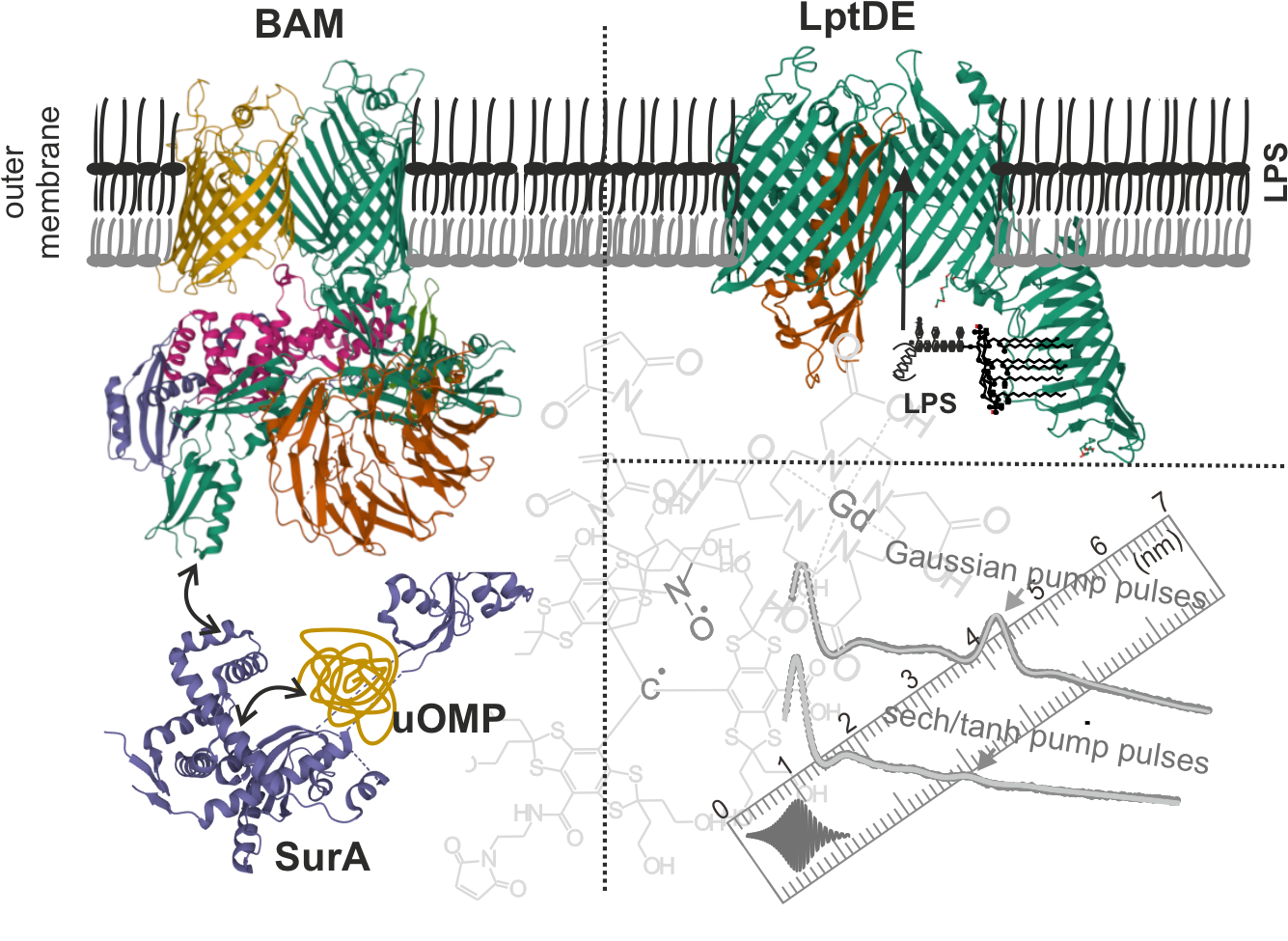
The cell envelope of Gram-negative bacteria consists of an inner membrane (IM) and an outer membrane (OM) that protects cells from harsh conditions. The OM is an asymmetric bilayer made up of phospholipids (PL) and lipopolysaccharides (LPS) and harbors numerous β-barrel proteins (outer membrane proteins, OMPs). The LptDE translocon mediates the final step of LPS insertion into the OM. Folding and insertion of unfolded OMPs from the periplasm into the OM is mediated by the β-barrel assembly machinery (BAM) with the assistance of periplasmic chaperones, primarily SurA. We will use in situ pulsed electron spin resonance (ESR) spectroscopy combined with other biophysical approaches to elucidate the structural and dynamical basis for outer membrane biogenesis. For SurA, its substrate-dependent conformational rearrangements and interaction with BAM complex will be investigated. For LptDE, conformational dynamics/heterogeneity of its key structural elements will be characterized in the native membrane environment in presence of LPS or the periplasmic LPS binding protein LptA. The binding and inhibition mode for the peptidomimetic antibiotic thanatin on LptDE will be investigated in a similar manner. Finally, we will develop new ESR approaches to study heterooligomeric OMP complexes in the native membranes employing different spin labels and unnatural amino acids.
P02: PROJECT-RELATED PUBLICATIONS
- Gopinath A, Rath T, Morgner N, Joseph B (2024) Lateral gating mechanism and plasticity of the ß-barrel assembly machinery complex in micelles and Escherichia coli. PNAS Nexus 3: pgae019
- Rudolph M, Tampé R, Joseph B (2023) Time-resolved Mn2+-NO and NO-NO distance measurements reveal that catalytic asymmetry regulates alternating access in an ABC transporter. Angew Chem Int Ed 62: e2023070
- Ketter S, Joseph B (2023) Gd(III)-trityl-nitroxide triple labeling and distance measurements in the heterooligomeric cobalamin transport complex in the native lipid bilayers. J Am Chem Soc 145: 2, 960-966
- Ketter S, Dajka M, Rogozhnikova O, Dobrynin S, Tormyshev VM, Bhagryanskaya EG, Joseph B (2022) In situ distance measurements in a membrane transporter using maleimide functionalized orthogonal spin labels and 5-pulse electron-electron double resonance spectroscopy. J Magn Reson Open, 10:100041
- Gopinath A and Joseph B (2021) Conformational flexibility of the protein insertase BamA in the native asymmetric bilayer elucidated with ESR spectroscopy. Angew Chem Int Ed 61: e202113448
- Ketter S, Gopinath A, Rogozhnikova O, Trukhin D, Tormyshev VM, Bhagryanskaya EG, Joseph B (2020) In situ labeling and distance measurements of membrane proteins in E. coli using Finland and OX063 trityl labels. Chem Eur J 27: 2299-2304
- Joseph B*, Jaumann EA, Barth K, Prisner TF, Cafiso DS (2019) In-situ observation of conformational dynamics and protein-ligand/substrate interactions in outer membrane proteins with DEER/PELDOR spectroscopy. Nat Prot 14: 2344-2369
- Chang YN, Jaumann EA, Reichel K, Hartmann J, Oliver D, Hummer G*, Joseph B* & Geertsma ER* (2019) Structural basis for functional interactions in dimers of SLC26 transporters. Nat Commun 10: 2032
- Barth K, Hank S, Spindler PE, Prisner TF, Tampé R, Joseph B (2017) Conformational coupling and trans-inhibition in the human antigen transporter ortholog TmrAB. J Am Chem Soc 140: 4527-4533
- Joseph B*, Tormyshev VM, Rogozhnikova OYu, Akhmetzyanov D, Bagryanskaya EG, Prisner TF* (2016) Selective high-resolution detection of membrane protein-ligand interaction in native membranes using trityl-nitroxide PELDOR. Angew Chem Int Ed 55: 11538-11542

