Project 12
Molecular dynamics simulations of membrane perforation machineries and the nuclear pore complex
Principle Investigator: Prof. Dr. Gerhard Hummer
Research Areas: Biophysics, Structural Biology
Summary
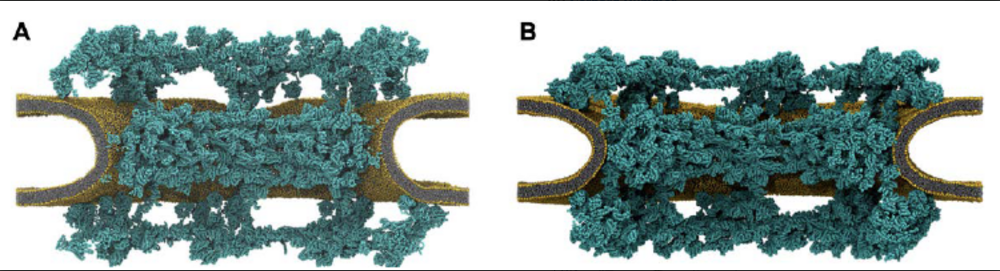
Complex machineries have evolved to mediate the transfer of large substrates across biological membranes, both into and out of cells and between cellular compartments. In P1, we propose to study key steps in the assembly and function of two archetypal pores in eukaryotes: (1) the ring-shaped cytolysin-like pores formed by gasdermin in inflammation, and (2) the nuclear pore complex (NPC) governing the transport between the nucleus and the cytosol. Aim 1 of this project is to resolve and characterize a key step in gasdermin-induced pore formation with the help of molecular dynamics simulations: the insertion of the beta-sheet structure into the membrane that then lines the wall of the pore. Gasdermin pores enable pro- inflammatory cytokine release and induce pyroptosis, a form of programmed cell death. Gasdermins recognize membranes in a lipid-specific manner, undergo major conformational changes upon membrane binding, assemble into large arcs and rings, and line the membrane pores with beta-barrel structures. To study gasdermin penetration into the membrane, we build on our studies of pore formation by bacterial cytolysins. To resolve the rare membrane insertion events, we will use path sampling methods. The simulations will allow us to test whether gasdermin penetration is cooperative, requires pre-ordered secondary structure, and involves specific lipids. As direct validation of key intermediates, we will work with P01 (Glaubitz) to characterize the structure of membrane-adhered and inserted peptides by solid- state nuclear magnetic resonance. Aim 2 focuses on the intracellular membrane pore that defines eukaryotes: the nuclear pore complex (NPC) governing the transport between the nucleus and the cytosol. Working closely with P15 (Beck, Lemke), we will assemble a detailed computational model of the NPC including the protein rings lining the pore, their anchors in the membrane of the nuclear envelope, the membrane itself, and the mesh of disordered FG repeats in the pore center. The NPC simulation model will allow us to test if the NPC is sensitive to mechanical perturbations of the nuclear envelope and if the patterned grafting of FG repeats contributes to NPC substrate selection and pore permeability. Our work builds on our integrative modeling studies of large membrane assemblies and of disordered proteins and protein condensates. The combination of molecular dynamics simulations with the electron tomography and fluorescence microscopy experiments of P15 promises to reveal nuclear transport at the molecular level and to give us a dynamic view of the NPC embedded in the nuclear envelope.

Prof. Dr. Gerhard Hummer
P12: PROJECT-RELATED PUBLICATIONS
- Johnson AG, Mayer ML, Schaefer SL, McNamara-Bordewick NK, Hummer G, Kranzusch PJ (2024) Structure and assembly of a bacterial gasdermin pore. Nature: preprint
- Wu D, Mehdipour A.R., Finke F, Goojani H.G., Groh R.R., Grund T.N, Reichhart T.M.B., Zimmermann R, Welsch S, Bald D, Shepherd M, Hummer G, Safarian S (2023) Dissecting the conformational complexity and mechanism of a prokaryotic heme transporter. Nat Chem Biol 19: 992-1003
- Yu M, Heidari M, Mikhaleva S, Tan P.S., Mingu S, Ruan H, Reinkemeier C.D., Obarska-Kosinska A, Siggel M, Beck M, Hummer G, Lemke E.A. (2023) Visualizing the disordered nuclear transport machinery in situ. Nature 617: 162-169
- Goretzki B, Wiedemann C, McCray B.A., Schäfer S.L., Jansen J, Tebbe F, Mitrovic S.A., Nöth J, Cabezudo A.C., Donohue J.K., Jeffries C.M., Steinchen W., Stengel F, Sumner C.J., Hummer G, Hellmich U.A. (2023) Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity. Nat Commun 14: 4165
- Jung H, Covino R, Arjun A, Leitpold C, Dellago C, Bolhuis P.G., Hummer G (2023) Machine-guided path sampling to discover mechanisms of molecular self-organization. Nat Comput Sci 3: 334-345
- Schäfer S, Hummer G (2022) Sublytic gasdermin-D pores captured in atomistic molecular simulations. eLife 11: e81432
- Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turoňová B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull MT, Hagen WJH, Hummer G, Kosinski J, Beck M (2022) AI-based structure prediction empowers integrative structural analysis of human nuclear pores. Science 376: eabm9506
- Sušac L, Vuong MT, Thomas C, von Bülow S, O’Brien-Ball C, Santos AM, Fernandes RA, Hummer G, Tampé R, Davis SJ (2022) Structure of a fully assembled tumor-specific T-cell receptor ligated by pMHC. Cell 185: 1-13
- Wu X, Siggel M, Ovchinnikov S, Mi W, Svetlov V, Nudler E, Liao M, Hummer G, Rapoport TA (2020) Structural basis of ERAD mediated by the Hrd1 ubiquitin ligase complex. Science 368: eaaz2449
- Turonová B, Sikora M, Schürmann C, Hagen WJH, Welsch S, Blanc FEC, von Bülow S, Gecht M, Bagola K, Hörner C, van Zandbergen G, Landry J, de Azevedo NTD, Mosalaganti S, Schwarz A, Covino R, Mühlebach MD, Hummer G, Krijnse Locker J, Beck M (2020) In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 370: 203-208
