Project 16
Dissecting the role of SND pathway components in ER membrane protein insertion
Principle Investigator: Dr. Melanie McDowell
Research Areas: Structural Biology, Biochemistry, Biophysics
Summary
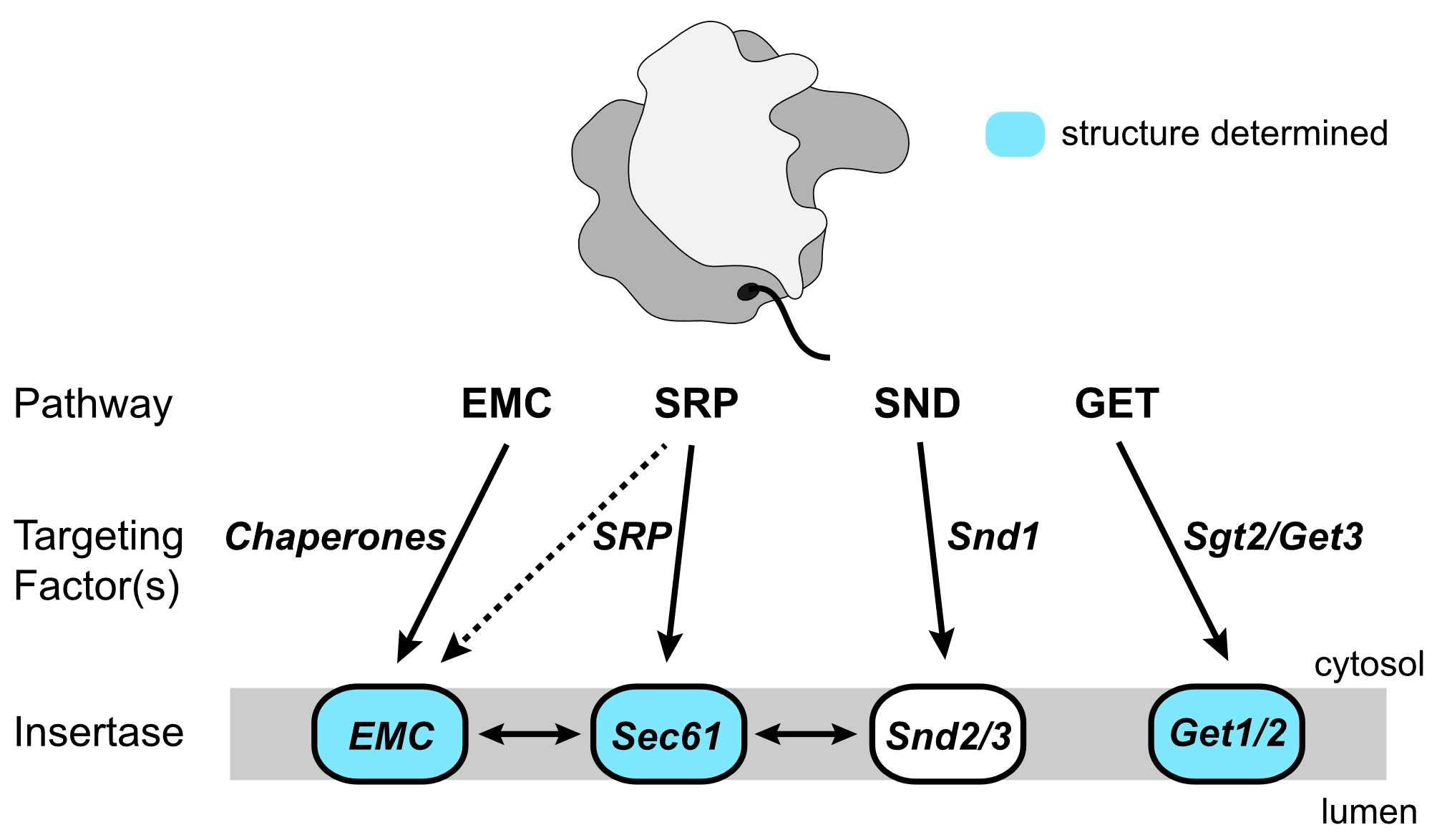
Most eukaryotic integral membrane proteins (IMPs) must be targeted to and inserted into the endoplasmic reticulum (ER) membrane after their translation by cytosolic ribosomes. As IMPs display a huge diversity in the quantity, relative positioning and biochemical properties of their transmembrane domains (TMDs), they require several distinct pathways for their ER targeting and insertion. Discovered in 2016, the signal recognition particle independent (SND) pathway specifically caters for IMP clients with their first TMD in a central position within the protein sequence, yet also provides an alternative route for more diverse IMP substrates when their canonical pathways are impaired. In Saccharomyces cerevisiae, three components of the SND pathway have been identified: the cytosolic protein Snd1 and the Snd2-Snd3 IMP complex. However, the characterization of the SND pathway is in its infancy, with little known about the interplay between these components and their precise roles. We propose to obtain a molecular-level understanding of the Snd proteins using a multi-faceted structural, biophysical and biochemical approach.
P16: PROJECT-RELATED PUBLICATIONS
- McDowell MA#, Heimes M#, Sinning I (2021) Structural and molecular mechanisms for membrane protein biogenesis by the Oxa1 superfamily. Nat Struct Mol Biol 28: 234-9
- McDowell MA*, Heimes M, Fiorentino F, Mehmood S, Farkas Á, Coy-Vergara J, Wu D, Reddy Bolla J, Schmid V, Heinze R, Wild K, Flemming D, Pfeffer S, Schwappach B, Robinson CV, Sinning I* (2020) Structural basis of tail-anchored membrane protein biogenesis by the GET insertase complex. Mol Cell 80: 72-86.e7
- McDowell MA#, Byrne AMP#, Mylona E, Johnson R, Sagfors A, Crepin VF, Lea SM, Frankel G (2019) The S. Typhi effector StoD is an E3/E4 ubiquitin ligase which binds K48- and K63-linked diubiquitin. Life Sci Alliance 2: e201800272
- McDowell MA, Marcoux J, McVicker G, Johnson S, Fong Y, Stevens R, Bowman LAH, Degiacomi MT, Yan J, Wise A, Friede ME, Benesch JLP, Deane JE, Tang CM, Robinson CV, Lea SM (2015) Characterisation of Shigella Spa33 and Thermotoga FliM/N reveals a new model for C-ring assembly in T3SS. Mol Microbiol 99: 749-66
- Abrusci P#, McDowell MA#, Lea SM, Johnson S (2014) Building a secreting nanomachine: a structural overview of the T3SS. Curr Opin Struct Biol 25: 111-7
- Berry J, Cehovin A, McDowell MA, Lea SM, Pelicic V (2013) Functional analysis of the interdependence between DNA uptake sequence and its cognate ComP receptor during natural transformation in Neisseria species. PLoS Genet 9: e1004014
- Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, Brady J, Baldwin GS, Lea SM, Matthews SJ, Pelicic V (2013) Specific DNA recognition mediated by a type IV pilin. Proc Natl Acad Sci USA 110: 3065-70
- Rollauer SE, Tarry MJ, Graham JE, Jääskeläinen M, Jäger F, Johnson S, Krehenbrink M, Liu SM, Lukey MJ, Marcoux J, McDowell MA, Rodriguez F, Roversi P, Stansfeld PJ, Robinson CV, Sansom MS, Palmer T, Högbom M, Berks BC, Lea SM (2012) Structure of the TatC core of the twin-arginine protein transport system. Nature 492: 210-4
- Zhong DM, Lefebre M, Kaur K, McDowell MA, Gdowski C, Jo S, Wang Y, Benedict SH, Lea SM, Galan JE, De Guzman RN (2012) The Salmonella type III secretion system inner rod protein PrgJ is partially folded. J Biol Chem 287: 25303-11
- McDowell MA, Johnson S, Deane JE, Cheung M, Roehrich AD, Blocker AJ, McDonnell JM, Lea SM (2011) Structural and functional studies on the N-terminal domain of the Shigella type III secretion protein MxiG. J Biol Chem 286: 30606-14

