Project 13
Probing multiprotein assemblies with native mass spectrometry
Principle Investigator: Prof. Dr. Nina Morgner
Research Areas: Native Mass Spectrometry
Summary
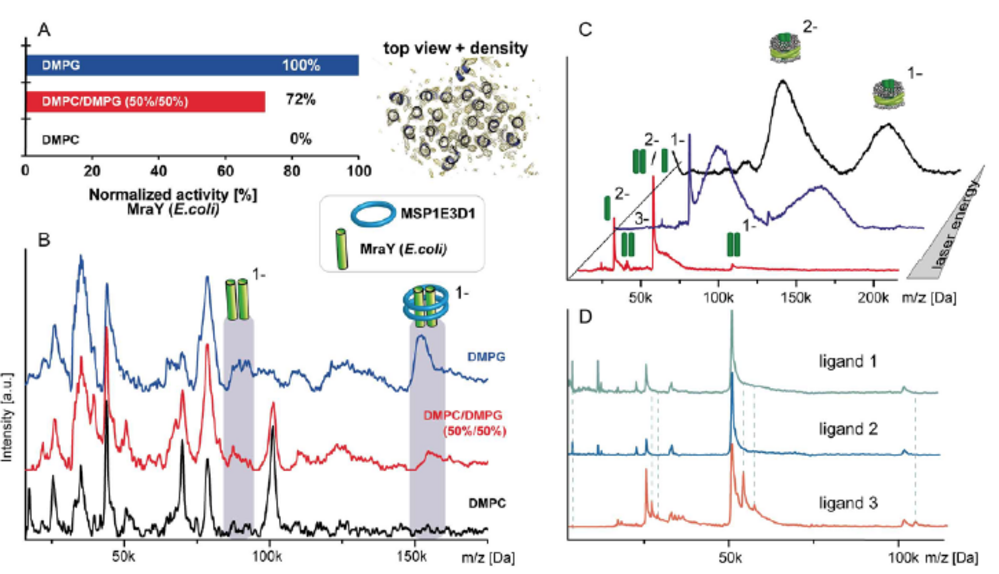
The correct (self-)assembly of molecular machines is of vital importance to ensure proper function of these multiprotein complexes. Analysis of this process and the necessary requirements, which allow unperturbed assembly into a complex’s functional state, is challenging. Native mass spectrometry allows us to analyze such protein assemblies regarding their stoichiometries, potential interaction partners as well as the assembly process itself. It is ideally suited to capture even the dynamic aspects of such processes, including intermediate steps in heterogeneous systems in dependence of well controlled environments or time dependencies. We want to understand the architecture and assembly mechanisms of multi- subunit machineries and assemblies, in the light of dependence on individual factors, such as the presence of nucleotides or chaperones in their reaction space, assembly factors needed for the assembly of different modules of a complex or the binding of specific substrates. In vitro assembly steps have to be probed individually and then correlated with results gained from similar experiments in more native environment. Especially for membrane proteins, native environments are still challenging to handle for many methods – we as well aim to push the boundaries regarding usable membrane mimics for native MS towards intact cellular membranes.

Prof. Dr. Nina Morgner
P13: PROJECT-RELATED PUBLICATIONS
- Young P, Hense G, Immer C, Wöhnert J, Morgner N (2020) LILBID laser dissociation curves: a mass spectrometry-based method for the quantitative assessment of dsDNA binding affinities. Sci Rep 10: 20398
- Lieblein T, Zangl R, Martin J, Hoffmann J, Hutchison MJ, Stark T, Stirnal E, Schrader T, Schwalbe H, Morgner N (2020) Structural rearrangement of amyloid-β upon inhibitor binding suppresses formation of Alzheimer’s disease related oligomers. eLife 9: e59306
- Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA (2019) Structure of the human TRPML2 ion channel extracytosolic/lumenal domain. Structure 27: 1246-57
- Peetz O, Hellwig N, Henrich E, Mezhyrova J, Dötsch V, Bernhard F, Morgner N (2019) LILBID and nESI: Different native mass spectrometry techniques as tools in structural biology. J Am Soc Mass Spectrom 30: 181-91
- Hellwig N, Peetz O, Ahdash Z, Tascón I, Booth PJ, Mikusevic V, Diskowski M, Politis A, Hellmich Y, Hänelt I, Reading E, Morgner N (2018) Native mass spectrometry goes more native: investigation of membrane protein complexes directly from SMALPs. Chem Commun 54: 13702-5
- Henrich E, Peetz O, Hein C, Laguerre A, Hoffmann B, Hoffmann J, Dötsch V, Bernhard F, Morgner N (2017) Analyzing native membrane protein assembly in nanodiscs by combined non-covalent mass spectrometry and synthetic biology. eLife 6: e20954
- Diskowski M, Mehdipour AR, Wunnicke D, Mills DJ, Mikusevic V, Barland N, Hoffmann J, Morgner N, Steinhoff HJ, Hummer G, Vonck J, Hänelt I (2017) Helical jackknives control the gates of the double-pore K+ uptake system KtrAB. eLife 6: e24303
- Angerer H, Schönborn S, Gorka J, Bahr U, Karas M, Wittig I, Heidler J, Hoffmann J, Morgner N, Zickermann V (2017) Acyl modification and binding of mitochondrial ACP to multiprotein complexes. Biochim Biophys Acta (BBA) – Mol Cell Res 1864: 1913-20.
- Brandt K, Muller DB, Hoffmann J, Langer JD, Brutschy B, Morgner N, Müller V (2016) Stoichiometry and deletion analyses of subunits in the heterotrimeric F-ATP synthase c ring from the acetogenic bacterium Acetobacterium woodii. FEBS J 283: 510-20.
- Bechara C, Nöll A, Morgner N, Degiacomi MT, Tampé R, Robinson CV (2015) A subset of annular lipids is linked to the flippase activity of an ABC transporter. Nat Chem 7: 255-62
