Project 18
Protein assemblies and machineries in antigen processing and ER quality control
Principle Investigator: Prof. Dr. Robert Tampé
Research Areas: Membrane biochemistry and biophysics, structural biology
Summary
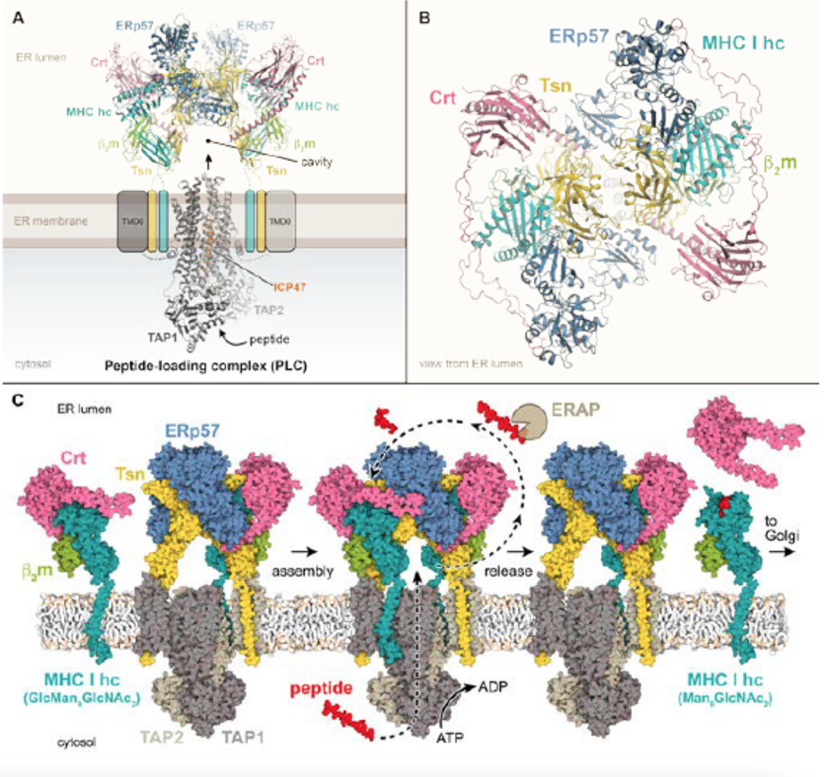
The adaptive immune system is built on sophisticated interplay of intracellular pathways and machineries of modularization, compartmentalization, and confinement. By monitoring peptide-MHC complexes, T-lymphocytes are able to identify and eliminate infected and cancerous cells. Stringent assembly of peptide-MHC I complexes occurs in a multitasking machinery, the MHC peptide-loading complex (PLC), in which the ABC transporter associated with antigen processing (TAP) shuttles proteasomal degradation products into the endoplasmic reticulum, resulting in the assembly and peptide loading of MHC I. Given their fundamental role within the immune system, these membrane-based assemblies are attractive targets for strategy development of viruses seeking to evade immune surveillance. Despite significant breakthroughs in elucidating the architecture of the multicomponent PLC and related quality control machineries, their mechanistic principles, dynamic assembly, and supramolecular organization remain to be adequately defined. By integrative biochemical and biophysical approaches, we will unravel how these immune service machineries operate temporally and spatially, and how viral factors come to modulate their function, supramolecular assembly and networking within membranes.

Prof. Dr. Robert Tampé
P18: PROJECT-RELATED PUBLICATIONS
- Barends M, Koller N, Schölz C, Durán V, Bošnjak B, Becker J, Döring M, Blees H, Förster R, Kalinke U, Tampé R (2023) Dynamic interactome of the MHC I peptide loading complex in human dendritic cells. Proc Natl Acad Sci USA 120: e2219790120
- Sagert L, Winter C, Ruppert I, Zehetmaier M, Thomas C, Tampé R (2023) The ER folding sensor UGGT1 acts on TAPBPR-chaperoned peptide-free MHC I. eLife 12: e85432
- Winter C, Domnick A, Cernova D, Tampé R (2022) Semisynthetic viral inhibitor for light control of the MHC I peptide loading complex. Angew Chem Int Ed 61: e202211826
- Müller IK, Winter C, Thomas C, Spaapen RM, Trowitzsch S, Tampé R (2022) Structure of an MHC I-tapasin-ERp57 editing complex defines chaperone promiscuity. Nat Commun 13: 5283
- Susac L, Young MT, Thomas C, von Bülow S, O’Brien-Ball C, Santos AM, Fernandes RA, Hummer G, Tampé R*, Davis SJ* (2022) Structure of a fully assembled tumor-specific T cell receptor ligated by pMHC. Cell 185: 3201-3213
- Domnick A, Winter C, Susac L, Hennecke L, Hensen M, Zitzmann N, Trowitzsch S, Thomas C, Tampé R (2022) Molecular basis of MHC I quality control in the peptide loading complex. Nat Commun 13: 4701
- Koller N, Höllthaler P, Barends M, Döring M, Spahn C, Duran V, Costa B, Becker J, Heilemann M, Klinke U, Tampé R (2022) Nanoscale organization of the MHC I peptide loading complex in human dendritic cells. Cell Mol Life Sci 79: 477
- Sánchez MF, Els-Heindl S, Beck-Sickinger AG, Wieneke R, Tampé R (2021) Photo- induced receptor-confinement drives ligand-independent GPCR signaling. Science 371: abb7657
- Sagert L, Hennig F, Thomas C, Tampé R (2020) A loop structure allow TAPBPR to exert its dual function as MHC I chaperon and peptide editor. eLife 9: e55326
- Hofmann S, Januliene D, Mehdipour AR, Thomas C, Stefan E, Brüchert S, Kuhn BT, Geertsma ER, Hummer G, Tampé R*, Moeller A* (2019) Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature 471: 580-3
