Project 17
Understanding nuclear pore complexity
Principle Investigator: Prof. Dr. Martin Beck, Prof. Dr. Edward A. Lemke
Research Areas: Chemical Biology, Imaging, Structural Biology
Summary
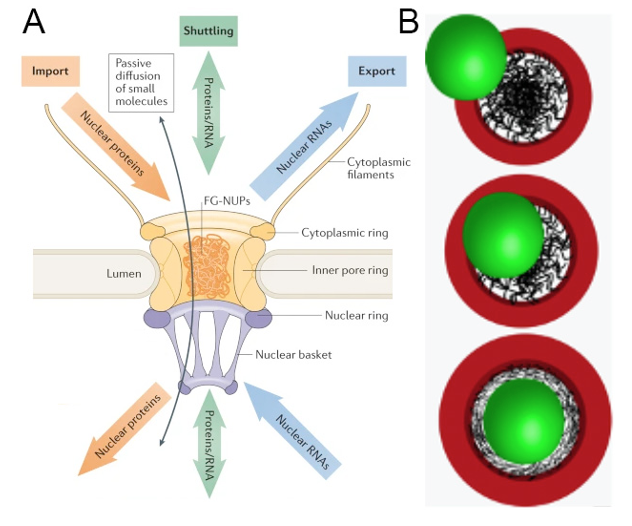
Nuclear pore complexes (NPCs) bridge the nuclear envelope and mediate nucleocytoplasmic exchange in a highly selective manner. NPCs are very intricate macromolecular assemblies made of about 1000 protein building blocks termed nucleoporins (Nups) that sum up to a molecular weight of ~110 MDa. About half of all Nups contain folded domains and form a cylindrical scaffold around a central channel. The other half is intrinsically disordered and rich in FG repeats. Intrinsically disordered Nups are grafted to the scaffold, thus lining the central channel. A subset of these intrinsically disordered Nups project through the scaffold towards the membrane. Recent structural work has revealed the positioning of most scaffold Nups within the assembly and consequently the grafting sites for FG Nups. The latter form a biophysical moiety that allows cargo complexes to pass, but remains inaccessible to inert molecules. However, the NPC is not a stiff channel. Intrinsically disordered Nups contain various interaction motifs in between flexible linkers and thus allow for conformational dynamics of the scaffold. NPC dilation and constriction by >20 nm that include a dynamic adaption of the nuclear membranes have been observed. Such large-scale conformational changes are critical for the transport of large cargo such as ribosomes and RNA, but also viruses. The NPC is potentially able to dilate by tens of nanometers, which moves the membrane embedding itself into the focus of understanding NPC function. However, an astonishing gap between structural and functional analysis of NPCs prevents a holistic understanding of the molecular mechanism of nucleocytoplasmic exchange and how conformational dynamics of NPCs, i.e. dilation and constriction, influences transport trajectories and the spatial distribution of Nups. Our hypothesis is that the biophysical properties of the nuclear envelope themselves actively contribute to NPC function, beyond merely anchoring the NPC. To address this challenge, we propose to develop a new integrated approach that systematically samples the missing information to understand nucleoporin dynamics. We will combine the expertise of the Lemke and Beck lab to develop a residue specific CLEM technology. This method will combine the power of site-specific fluorescence labelling to study Nups with the power of in situ cryo electron tomography to monitor the conformation of the NPC. Guided by computational model building of the Hummer lab, we will aim to reveal those parts of the NPC which are currently elusive to either of the three methods in context of different biological states, such as cellular stress or for specific cargos. During the first funding period, we will focus on technology development to correlate large cargo transport with NPC dilation to understand how Nup function projects into the nuclear envelope and vice versa. Iterative exchange of information between computational model building and experimental measurements should ultimately lead us to a time-resolved view and an understanding of how the membrane breathes together with the NPC and how substantial spatial rearrangements in the NPC structure and dynamics are facilitated or constrained by the membrane environments across the wide range of distinct physiological activities. As a long-term goal, we want to understand the complexity of nuclear transport at the molecular level within the context of whole nuclear envelope architecture and design, and move beyond a static view that treats the NPC as rigid and purely proteinacious transport channel. Our aim to illuminate this fundamental process of life is driven also by its implications for nuclear transport associated disease states such as neurodegenerative and genetic diseases.
P17: PROJECT-RELATED PUBLICATIONS
- Hoffmann PC, Kim H, Obarska-Kosinska A, Kreysing JP, Andino-Frydman E, Cruz-Leon S, Margiotta E, Cernikova L, Kosinski J, Turonova B, Hummer G*, Beck M* (2025) Nuclear pore permeability and fluid flow are modulated by its dilation state. Mol Cell 85, 537-554
- Kreysing JP, Heidari M, Zila V, Cruz-Leon S, Obarska-Kosinska A, Laketa V, Rohleder L, Welsch S, Köfinger J, Turonová B, Hummer G*, Kräusslich HG*, Beck M* (2025) Passage of the HIV capsid cracks the nuclear pore. Cell 188, 930-943
- Taniguchi R, Omiacki C, Kreysing JP, Zila V, Zimmerli CE, Bohm S, Turonová B, Kräusslich HG, Doye V* Beck M* (2025) Nuclear pores safeguard the integrity of the nuclear envelope. Nat Cell Biol 27: 762-775
- Yu M, Heidari M, Mikhaleva S, Tan PS, Mingu S, Ruan H, Reinkemeier CD, Obarska-Kosinska A, Siggel M, Beck M, Hummer G, Lemke EA (2023) Visualizing the disordered nuclear transport machinery in situ. Nature 617: 162-169
- Zila V, Margiotta E, Turoňová B, Müller TG, Zimmerli CE, Mattei S, Allegretti M, Börner K, Rada J, Müller B, Lusic M, Kräusslich HG*, Beck, M* (2021) Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 184: 1032-46
- Zimmerli CE, Allegretti M, Rantos V, Goetz SK, Obarska-Kosinska A, Zagoriy I, Halavatyi A, Hummer G, Mahamid J, Kosinski J*, Beck M* (2020) Nuclear pores dilate and constrict in cellulo. Science 374: eabd9776
- Paci G, Zheng T, Caria J, Zilman A*, Lemke EA* (2020) Molecular determinants of large cargo transport into the nucleus. eLife 9: e55963
- Allegretti M, Zimmerli CE, Rantos V, Wilfling F, Ronchi P, Fung HKH, Lee CW, Hagen W, Turoňová B, Karius K, Börmel M, Zhang X, Müller C, Schwab Y, Mahamid J, Pfander B*, Kosinski J*, Beck M* (2020) In cell architecture of the nuclear pore complex and snapshots of its turnover. Nature 586: 796–800
- Reinkemeier CD, Estrada Girona G, Lemke EA (2019) Designer membraneless organelles enable codon reassignment of selected mRNAs in eukaryotes. Science 363: eaaw2644
- Hampoelz B, Schwarz A, Ronchi P, Bragulat-Teixidor H, Tischer C, Gaspar I, Ephrussi A, Schwab Y, Beck M (2019) Nuclear Pores Assemble from Nucleoporin Condensates During Oogenesis. Cell 179: 671-86


